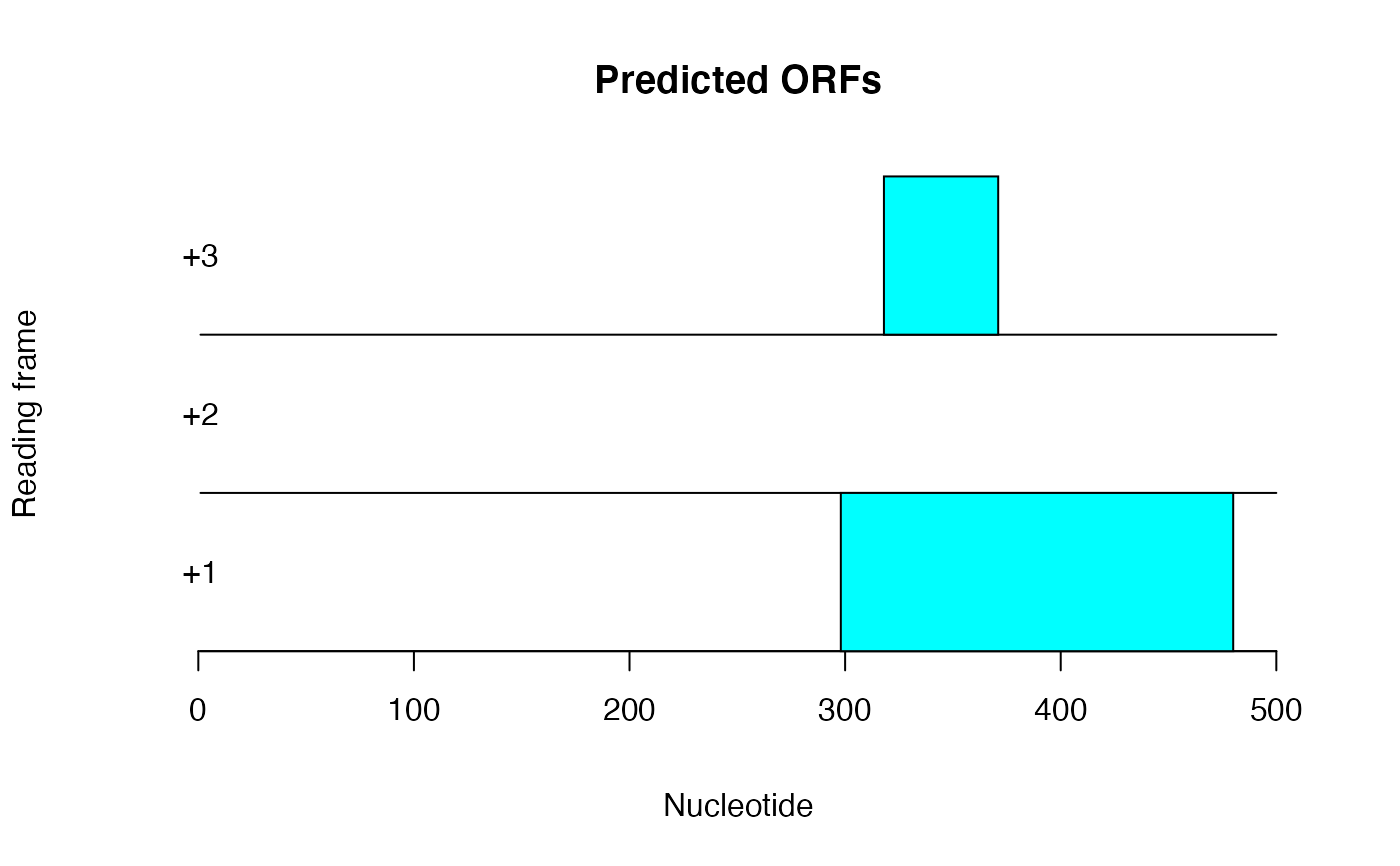Find and plot potential ORFs
plot_ORFs_in_seq.RdBy Coghlan (2011) A little book of R for bioinformatics. https://a-little-book-of-r-for-bioinformatics.readthedocs.io/en/latest/. This function calls find_start_stop_ORFs() thenm builds a plot of the output. Predictions are based on the standard genetic code.
plot_ORFs_in_seq(sq)
Arguments
| sq | DNA sequence |
|---|
Examples
# From Coglan 2011 ## Toy example s1 <- "aaaatgcagtaacccatgccc" plot_ORFs_in_seq(s1)